In this NewsFlash, tailor made proteins that bind to 'flu viruses, the largest gathering of whale sharks in the world, and how induced stem cells may be rejected, even by genetically identical animals. Plus, how a laser technique has shed new light on a common process that leads to cancer.
In this episode
00:27 - Bespoke Proteins Bind ‘Flu
Bespoke Proteins Bind ‘Flu
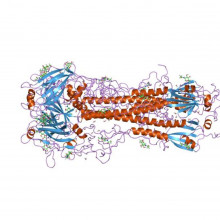
Researchers in Seattle and Washington have solved an "enormous jigsaw puzzle" to design two novel proteins that bind to a protein found in influenza viruses, proving that computer designed proteins are feasible and could form the basis of new drugs and biosensors. They report on the work in the journal Science this week.
Protein-protein interactions are a common biochemical process, important in a great many biological systems, and are controlled by their molecular structure. For two proteins to bind, they need complimentary shapes that can fit together like a lock and key, with no overlap and little empty space.
 Sarel J. Fleishman at the University of Washington, along with colleagues in California, used cutting edge software and over 100,000 hours of parallel computing time to design new proteins that would bind to the tail of a common virus protein, Hemagglutinin, from the 1918 H1N1 'flu strain. This protein helps the virus to invade cells, and is found in many strains of influenza. Most natural antibodies bind to the variable head of the protein, leaving room for the virus to evolve immunity, but the tail is highly conserved across strains, making it an excellent target for attack.
Sarel J. Fleishman at the University of Washington, along with colleagues in California, used cutting edge software and over 100,000 hours of parallel computing time to design new proteins that would bind to the tail of a common virus protein, Hemagglutinin, from the 1918 H1N1 'flu strain. This protein helps the virus to invade cells, and is found in many strains of influenza. Most natural antibodies bind to the variable head of the protein, leaving room for the virus to evolve immunity, but the tail is highly conserved across strains, making it an excellent target for attack.
To design the protein, they first identified hot-spots of interaction on the virus protein. These are areas where hydrogen bonding and electrostatic interactions allow for low-energy bonding of complimentary structures. With a map of these hot-spots, they set about putting the jigsaw pieces together and designing a structure that would compliment, and therefore bind to, the protein.
Candidate proteins were tested using a highly efficient yeast-based assay, and two of the designed proteins, HB36 and HB80, were shown to bind well with the 1918 H1N1, as well as others strains of H1N1 and H5N1. One of the proteins, HB80, also inhibits protein changes important in infection.
Although by no means perfect, and there will be many hurdles to cross before designed proteins become useful medications, this is an excellent example of successful computer-aided-protein design, a technique that could be instrumental in future diagnostics and treatment.

03:23 - Uncovering a whale shark extravaganza
Uncovering a whale shark extravaganza
Scientists in Mexico have discovered the largest mass gathering of whale sharks in the world.
 These gentle giants can grow up to around 12 metres or 40 feet in length, which means spotting just one of them as they cruise the oceans is an unforgettable experience. Imagine what it must be like spotting a gang of more than 400 whale sharks?
These gentle giants can grow up to around 12 metres or 40 feet in length, which means spotting just one of them as they cruise the oceans is an unforgettable experience. Imagine what it must be like spotting a gang of more than 400 whale sharks?
That's what a team of researchers from Mexico and the US did back in the summer of 2006, when the huge aggregation site, which they've named Afuera, was spotted from a plane flying off the coast of the Yucatan Peninsular in the Caribbean Sea. Since then they've been back each year to carry out studies from the air and in the water to try and figure out what's going on. And in 2009, they spotted the largest aggregation of 420 sharks in an area covering just 12 square km.
The big question is, why do whale sharks do it?
When any animals group together en masse there are really only two possible explanations: sex or food. And in the case of the whale sharks at Afuera, and elsewhere, where other, smaller aggregations form, it turns out it's the latter.
Whale sharks gather at Ningaloo Reef in Western Australia to feed when the coral reefs undergo mass spawning. At another aggregation in Belize, whale sharks are after the eggs of Dog snappers that also spawn in aggregations.
At Afuera, the research team sieved the sea for plankton and found that is was awash with fish eggs. And DNA barcoding revealed that they're a type of tuna called the little tunny. These eggs come packed with fats, making them superb whale shark food. And it's thought that the reason the tuna show up and spawn in this spot is thanks to the upwelling along the coast that injects a pulse of nutrients into the ecosystem.
There's another, smaller aggregation close to Afuera that's been known about for a few years and has recently been protected by the Mexican government. That one already draws in flocks of tourists who - quite understandably - are keen to swim with the biggest sharks in the ocean.
But researchers are worried that this could cause problems with collisions between sharks and high-speed boats. So they're calling for swift action to protect the animals in this extraordinary natural event, and so hopefully they'll still be there for generations to come.
Find out more
An unprecedented aggregation of whale sharks, Rhincodon typus, in Mexican Coastal waters of the Caribbean Sea
06:22 - Lasers Identify Key Molecular Structure in Tumours
Lasers Identify Key Molecular Structure in Tumours
Dr Marisa Martin-Fernandez; STFC’s Central Laser Facility & Dr Martyn Winn; Computational Science and Engineering Department
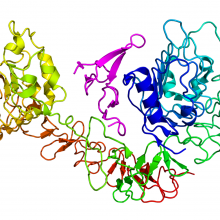
Ben - Also in the news this week, a laser technique has exposed the previously unknown molecular shape of Epidermal Growth Factor Receptors or EGFRs which are known to be involved in the development of cancer. These are found on the surface of the vast majority of human cells as Dr. Marisa Martin-Fernandez, a scientist based at the STFC's Central Laser Facility in Harwell explained...
Marisa - The role of this receptor is to bind molecules which are in the bloodstream and produce signals which are transduced to the inside of each cell and to give instructions to the cell machinery to grow, divide, die, differentiate. So it's kind of the core of how to promote cell behaviour. These are the orchestrators, these type of receptors orchestrate what cells are going to do within a multicellular body and there is a lot that is known about what happens in this signalling process that leads to cellular growth and when it goes wrong and leads to the growth of tumours. In fact, most human tumours have a fault in the signalling of this receptor.
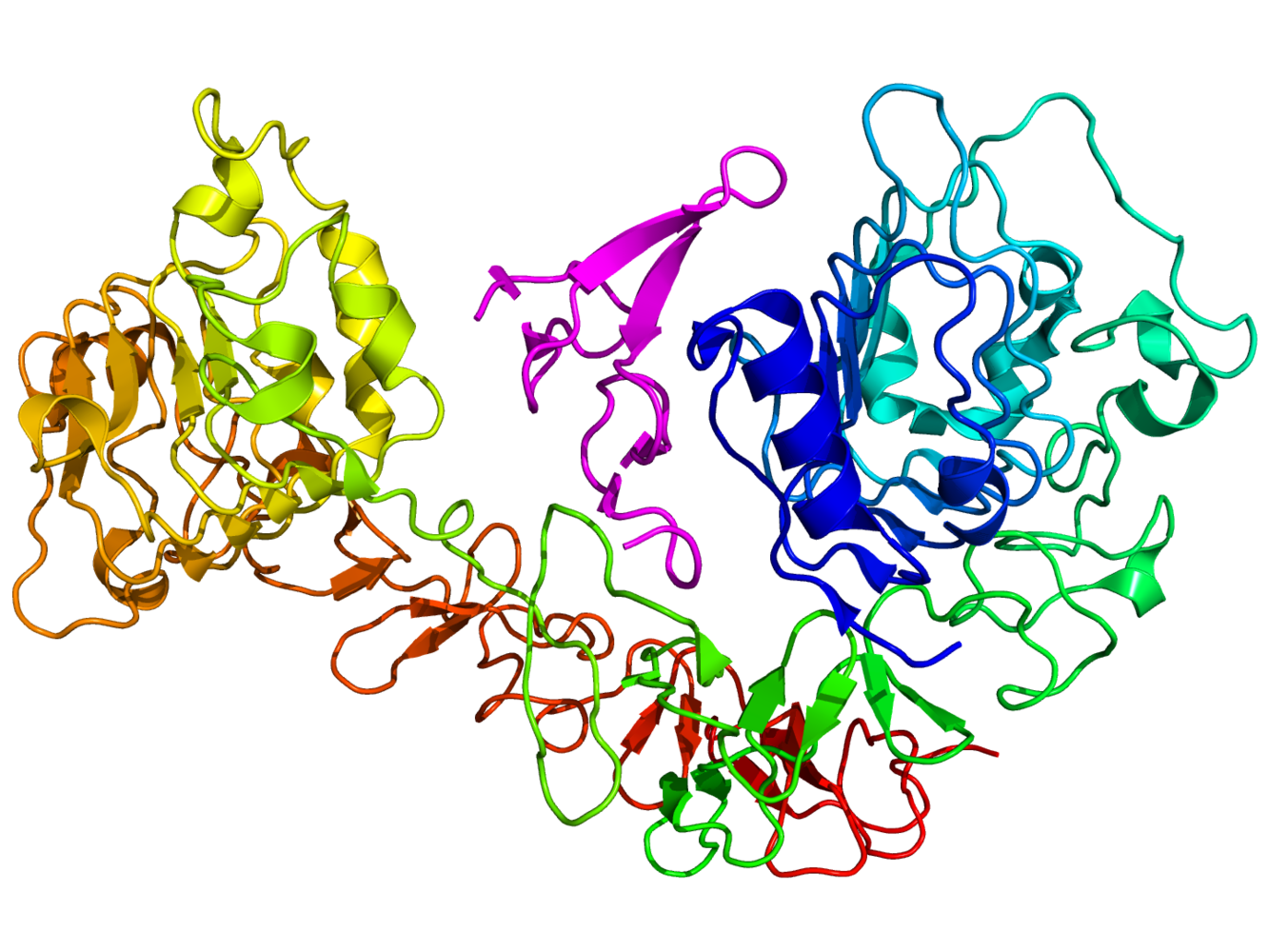 Ben - Current drugs that target EGFRs do so non-discriminately, blocking activation to halt cell growth. However, in doing so, they may block other essential cell maintenance and this could lead to the body adopting other chemical pathways to achieve the same goal. This side steps the cancer treatment and allows the tumour to grow once more. To find out more about the precise interactions between EGFRs and signalling molecules, we need a good understanding of their structure. To do this, Marisa's team using novel laser technique called fluorescence resonance energy transfer...
Ben - Current drugs that target EGFRs do so non-discriminately, blocking activation to halt cell growth. However, in doing so, they may block other essential cell maintenance and this could lead to the body adopting other chemical pathways to achieve the same goal. This side steps the cancer treatment and allows the tumour to grow once more. To find out more about the precise interactions between EGFRs and signalling molecules, we need a good understanding of their structure. To do this, Marisa's team using novel laser technique called fluorescence resonance energy transfer...
Marisa - So what we do is we put a label in a position in the receptor that we can control, which is a molecule that emits fluorescence, and we put another label on the cell surface, and then we excite one of the molecules in the receptor. Depending on how close it is to the surface, the characteristics of the fluorescence emission are different and from that information, we can actually get nanometer distances.
Ben - Knowing the distances between the receptor and the other parts of the cell can help to simulate exactly what structure the receptor will take in situ. This job is taken on by experts such as Dr. Martyn Winn of the STFC's Computational Science and Engineering Department at the Daresbury laboratory...
Martyn - Our starting points are atomic structures that you get from crystallography. These are experiments that would take place on Diamond Light Source, for example, and they give you very detailed atomic structures of the proteins that are involved. The drawback of that is they're static. They don't move around like they do in real life. Also, that means that the proteins have been taken out of the cell, taken out of their natural context and put into a crystal. So you have very detailed information but it's not necessarily relevant to what's happening in the cell. So what we would do in a simulation is to take that detailed structure, set up a simulation which mimics the cell environment and then see what happens when it's put into the cellular context.
Ben - When Marisa's laser data was combined in a simulation with the known atomic structure of EGFRs, the resulting shape was new, unexpected, but surprisingly similar to a structure found in Drosophila, the fruit fly.
Martyn - So what you often see from crystallography is very symmetric structures. The crystal environment is a very symmetric environment and everything is nice and well ordered. What we saw when we did our simulation was that the molecule actually becomes asymmetric. When we first did this, this was something that had not been seen before and slightly heretic. People love symmetry. Symmetry is beautiful, but our simulations were showing that the molecule became asymmetric in order to explain Marisa's experiments.
Marisa - And then the amazing thing is that when we look at the molecular shape, it looked nearly identical to the molecular shape of the Drosophila receptor. And then when we identified the shape, it was virtually identical. It was so similar, it was actually uncanny. We were amazed, when we looked at the structure that we identified, and it looked like the Drosophila receptor, we knew we were onto something.
Ben - So, knowing this particular structure could help lead to better therapeutics and the technique itself could help move forward into personalised medicine.
Marisa - Given this new information, we could actually consider new antibody therapeutics that could block one of the other version of the receptor and allow the other signals to be transduced. So, in a way, they will be less invasive therapeutics that might actually reduce side effects, and make sure that, for example, the body is not deprived of a fundamental function that this receptor covers.
Martyn - And I think we're at the level of very basic science and it's a long road towards drug development, that's true, but I think one of the things that they want to do is to develop treatments that are specific to patients. So you can analyse the DNA from particular patients to see if they have particular mutations. It might be that you can also analyse the tumours of patients to see what the confirmation of the receptors are and then that might shed light on whether a particular antibody treatment is likely to work or not. That their interest, actually in implying this to patient-specific treatments.
Ben - That was Martyn Winn from the Daresbury Lab and Marisa Martin-Fernandez from the Central Laser Facility in Harwell. You can find that paper in the journal Molecular and Cellular Biology.
12:49 - Immune Roadblock to Stem Cell Treatment
Immune Roadblock to Stem Cell Treatment
Stem cells may be rejected by the animals they first came from, according to research published in the journal Nature this week. This could be a huge stumbling block in the use of induced stem cells for therapy.
Induced Pluripotent stem cells, or iPSCs, are created by taking normal cells from an animal, then exposing them to factors that could allow them to differentiate into any of the body's cells, much like embryonic stem cells. As they are genetically identical to the animal, it's been assumed that they would not cause an immune response, and therefore avoid rejection.
 Now, Tongbiao Zhao and colleagues at the University of California, San Diego, have tested that assumption, and found that, at least for 2 of the methods of inducing pluripotency, it doesn't hold true.
Now, Tongbiao Zhao and colleagues at the University of California, San Diego, have tested that assumption, and found that, at least for 2 of the methods of inducing pluripotency, it doesn't hold true.
The team took mouse fibroblasts, cells that make up the structural component of skin, and used two different techniques to turn them into iPSCs before transplanting them into genetically identical mice. They did the same with embryonic stem cells for comparison.
The results were startling. The embryonic stem cells were mostly able to grow and divide within the mice, but the induced stem cells triggered an immune attack, growing to far smaller sizes and showing damage typical of immune rejection. Induced cells transplanted into immune deficient mice were able to grow and divide as normal.
Genetic comparisons showed certain genes were over expressed in the induced compared to embryonic stem cells, and subsequent testing showed that this is most likely to be responsible for inducing a T-cell mediated immune response.
This is a stumbling block for induced stem cell therapies, and shows that we need to ensure that cells created in this way are not just genetically identical to embryonic stem cells, but that we also need to account for epigenetic factors such as gene expression.
Related Content
- Previous Does loud music annoy whales?
- Next Wet But Not Wild - Farming Fish










Comments
Add a comment